
Research
OVERVIEW
- Hospitals today are collecting an immense amount of patient data (e.g., images, lab tests, vital sign measurements). Despite the fact that health data are messy and often incomplete, these data are useful and can help improve patient care. To this end, we have pioneered work in developing machine learning (ML) and artificial intelligence techniques (AI) inspired by problems in healthcare (e.g., predicting patient outcomes like infections).
In collaboration with 30+ clinicians and domain experts, we have identified areas with potential for high clinical impact and methodological impact. Through extensive preliminary work dedicated to understanding the clinical problem and the dataset that will be used, we formalize problems and develop new approaches. Our methodological contributions cluster into the following technical thrusts, driven primarily by four questions related to "who," "why," "what," and "how".
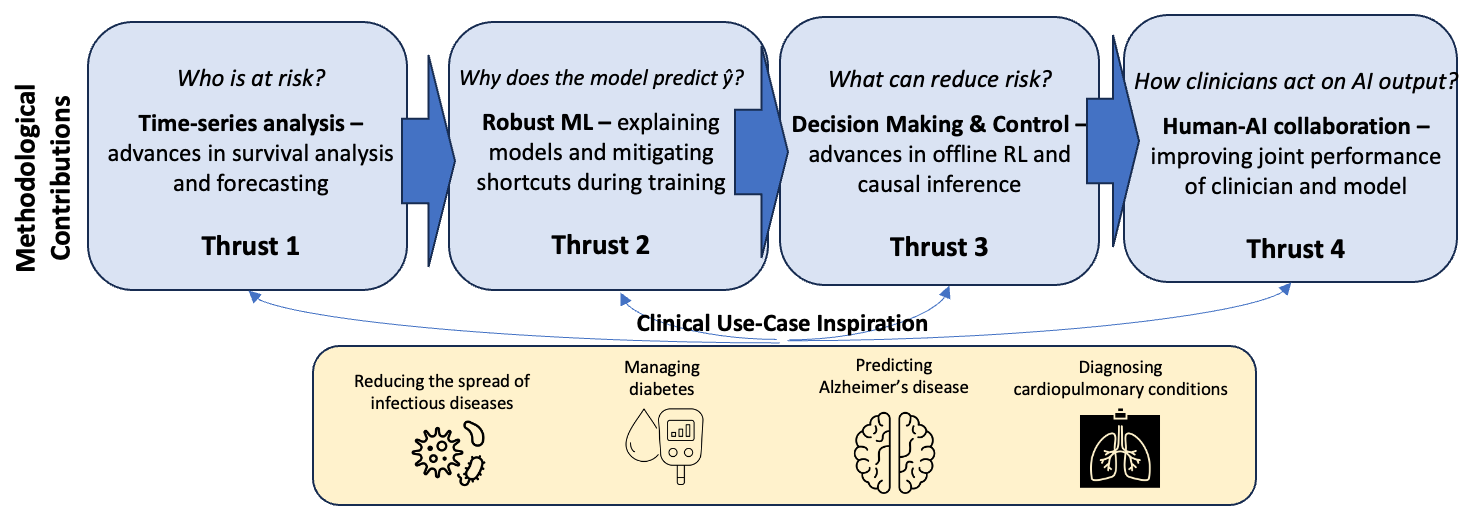
METHODOLOGICAL CONTRIBUTIONS
Thrust 1: Times-series Analysis - We work on approaches for predicting outcomes from time-series data and time-series forecasting. Modeling health data using the trajectory of a patient instead of snapshots led to new challenges and pitfalls highlighted by my group [ AMIA’17 KDDM Best Paper], which continues to be used as a guide in setting up new prediction tasks. While much emphasis is often placed on the learning architecture/objective in ML, >90% of the work is preprocessing. To this end, we developed an open-source software package, “FIDDLE,” that incorporates best practices from the literature with the goal of streamlining the processing of time-series clinical data [JAMIA ‘20]. This tool has been used over 100 times since its initial publication. In time-series analysis, the output can also vary over time. To this end we have made significant contributions to survival analysis [AAAI ‘21a, AAAI ‘21b, ICDM ’24] and time-series forecasting [KDD ‘18, AAAI ‘23], winning challenges [KDH @ ECAI ‘20].
Thrust 2: Robust ML - Deep learning approaches while powerful are prone to latching on to spurious correlations or ‘shortcuts’ that hold in the training dataset but fail to generalize. E.g., if the sickest patients in a hospital are all located in a unit where a specific chest x-ray machine is used, a model could pick up on artifacts in the training data associated with the machine used to capture the chest x-ray, rather than clinically-relevant radiological findings. While the model has learned an accurate association in the data it is unlikely to generalize across institutions or even across time, within the same hospital. We have highlighted the dangers of shortcuts in health datasets [MLHC ‘20], and have worked on mitigating shortcuts during model training to increase model robustness [KDD‘18, NeurIPS ’22a, KDD ‘25]. Systematic under-testing of subpopulations can lead models to wrongly associate them with lower risk; we introduced disparate censorship to study differences in testing rates across patient groups as a source of bias [MLHC ‘22, PLOS ’24], and proposed a solution [ICML’24].
Thrust 3: Decision Making & Control - With the goal of improving the actionability of AI models in health, I have focused on tackling challenges in learning treatment policies using causal inference and offline reinforcement learning (RL). My work provides theoretical, as well as practical, foundations for human-in-the-loop decision making, in which humans (e.g., clinicians, patients) can incorporate additional knowledge (e.g., side effects, patient preference) when selecting among near equivalent actions or treatments. Our foundational advancements resulted in a novel model-free algorithm for learning “set-valued policies” (SVPs), which returns a set of near-equivalent actions rather than a single optimal action [ICML ‘20]. Recognizing that many problems in healthcare result in exponential action spaces when treatments are considered in combination, we also developed an approach for sample-efficient offline RL with factored action spaces [NeurIPS ’22b] and a new semi-offline paradigm for model policy evaluation [NeurIPS ‘23]. We have worked to increase the rigor of offline RL in healthcare with our work in model selection for offline RL [MLHC ‘21a] that has been cited >100 times. This work has also led to contributions related to “decision-focused” learning [AISTATS ‘24, JAMIA ‘25].
Thrust 4: Human-AI collaboration - As we move towards integrating these tools into clinical workflows, we have started to examine how clinicians interacted with the output of an AI model and specifically how it influences decisions. We showed how an incorrect model can harm clinician accuracy and lead to poor treatment decisions, and that image-based explanations do nothing to mitigate that harm [JAMA ‘23]. This finding is particularly important given that explanations were emphasized by the White House blueprint for an AI Bill of Rights. We continue to study the impact of selective prediction, and how hiding potentially unreliable model predictions may reduce automation bias.
Preprints/Publications/Presentations
2025
- Shengpu Tang, Stephanie Shepard, Rebekah Clark, Erkin Ötleş, Chidimma Udegbunam, Josh Tran, Melinda Seiler, Justin Ortwine, Akbar K. Waljee, Jerod Nagel, Sarah L. Krein, Jacob E. Kurlander, Paul J. Grant, Jihoon Baang, Anastasia Wasylyshyn, Krishna Rao, Jenna Wiens Guiding Clostridioides difficile Infection Prevention Efforts in a Hospital Setting With AI , Journal of the American Medical Informatics Association, 2025.
- Fahad Kamran, Donna Tjandra, Thomas S. Valley, Hallie C. Prescott, Nigam H. Shah, Vincent X. Liu, Eric Horvitz, and Jenna Wiens Reformulating patient stratification for targeting interventions by accounting for severity of downstream outcomes resulting from disease onset: a case study in sepsis , Journal of the American Medical Informatics Association, 2025.
- Shalmali Joshi, Iñigo Urteaga, Wouter AC van Amsterdam, George Hripcsak, Pierre Elias, Benjamin Recht, Noémie Elhadad, James Fackler, Mark P Sendak, Jenna Wiens, Kaivalya Deshpande, Yoav Wald, Madalina Fiterau, Zachary Lipton, Daniel Malinsky, Madhur Nayan, Hongseok Namkoong, Soojin Park, Julia E Vogt, and Rajesh Ranganath AI as an intervention: improving clinical outcomes relies on a causal approach to AI development and validation , Journal of the American Medical Informatics Association, 2025.
- Michael Ito, Jiong Zhu, Dexiong Chen, Danai Koutra and Jenna Wiens Leaning Laplacian Positional Encodings for Heterophilous Graphs , AISTATS, 2025.
- Michael Ito, Danai Koutra and Jenna Wiens Understanding GNNs and Homophily in Dyname Node Classification , AISTATS, 2025.
2024
- Trenton Chang, Mark Nuppnau, Ying He, Keith E. Kocher, Thomas S. Valley, Michael W. Sjoding, and Jenna Wiens Racial differences in laboratory testing as a potential mechanism for bias in AI: a matched cohort analysis in emergency department visits , PLOS Global Public Health, 2024.
- Harry Rubin-Falcone, Joyce Lee, and Jenna Wiens Learning control-ready forecaster for Blood Glucose Management , Computers in Biology and Medicine, 2024.
- Jung Min Lee, Rodica Pop-Busui, Jasper Fleischer, and Jenna Wiens Shortcomings in the Evaluation of Blood Glucose Forecasting , IEEE Transactions on Biomedical Engineering, 2024.
- Rohan Khera, Evangelos K Oikonomou, Girish N Nadkarni, Jessica R Morley, Jenna Wiens, Atul J Butte, and Eric J Topol Transforming Cardiovascular Care With Artificial Intelligence: From Discovery to Practice: JACC State-of-the-Art Review , Journal of the American College of Cardiology, 2024.
- Jenna Wiens, Kayte Spector-Bagdady, and Bhramar Mukherjee Toward Realizing the Promise of AI in Precision Health Across the Spectrum of Care , Annual Review of Genomics and Human Genetics, 2024.
- Meera Krishnamoorthy, Michael Sjoding, and Jenna Wiens Off-label use of AI models in Healthcare , Nature Medicine, 2024.
- Rohan Khera and Jenna Wiens Summertime for Cardiovascular AI , Circulation: Cardiovascular Quality and Outcomes, 2024.
- Chase Irwin, Donna Tjandra,Chengcheng Hu, Vinod Aggarwal, Amanda Lienau, Bruno Giordani, Jenna Wiens, Raymond Q Migrino Predicting 5-year dementia conversion in veterans with mild cognitive impairment , Alzheimer’s & Dementia: Diagnosis, Assessment & Disease Monitoring, 2024.
- Fahad Kamran, Donna Tjandra, Andrew Heiler, Jessica Virzi, Karandeep Singh, Jessie E King, Thomas S Valley, and Jenna Wiens Evaluation of Sepsis Prediction Models before Onset of Treatment , NEJM AI, 2024.
- Trenton Chang, Lindsay Warrenburg Sae-Hwan Park, Ravid B Parikh, Maggie Makar and Jenna Wiens Who’s Gaming the System? A Causally-Motivated Approach to Detecting Strategic Adaptation , NeurIPS, 2024.
- Sarah Jabbour, Gregory Kondas, Ella Kazerooni, Michael Sjoding, David Fouhey, and Jenna Wiens DEPICT: Diffusion-Enabled Permutation Importance for Image Classification Tasks , ECCV, 2024.
- Meera Krishnamoorthy and Jenna Wiens Multiple Instance Learning with Absolute Position Information , CHIL, 2024.
- Trenton Chang, and Jenna Wiens From Biased Selective Labels to Pseudo-Labels: An Expectation-Maximization Framework for Learning from Biased Decisions , ICML, 2024.
- Fahad Kamran, Maggie Makar, and Jenna Wiens Learning to Rank Optimal Treatment Allocation Under Resource Constraints , AISTATS, 2024.
- Trenton Chang, Jenna Wiens, Tobias Schnabel, and Adith Swaminathan Measuring Steerability in Large Language Models , NeurIPS Safe Generative AI Workshops, 2024.
2023
- Sarah Jabbour, David Fouhey, Stephanie Shepard, Thomas S Valley, Ella A Kazerooni, Nikola Banovic, Jenna Wiens and Michael Sjoding Measuring the impact of AI in the diagnosis of hospitalized patients: a randomized clinical vignette survey study , JAMA 330 (23), 2275-2284, 2023.
- Meghana Kamineni, Erkin Otles, Jeeheh Oh, Krishna Rao, Vincent B Young, Benjamin Y Li, Lauren R West, David C Hooper, Eric S Shenoy, John Guttag, Jenna Wiens, and Maggie Makar Prospective evaluation of data-driven models to predict daily risk of Clostridioides difficile infection at 2 large academic health centers , Infection Control & Hospital Epidemiology, 44(7) 2023.
- Erkin Ötles, Emily A Balczewski, Micah Keidan, Jeeheh Oh, Alieysa Patel, Vincent B Young, Krishna Rao, and Jenna Wiens Clostridioides difficile infection surveillance in intensive care units and oncology wards using machine learning , Infection Control & Hospital Epidemiology, 2023.
- Courtney Armour, Kelly Sovacool, William Close, Begum Topcuoglu, Jenna Wiens, and Patrick Schloss Machine learning classification by fitting amplicon sequences to existing OTUs , MSphere(8) 5 2023.
- Westyn Branch-Elliman, Alexander J Sundermann, Jenna Wiens, and Erica S Shenoy The future of automated infection detection: innovation to transform practice , Antimicrobial Stewardship & Healthcare Epidemiology, 3(1), 2023.
- Westyn Branch-Elliman, Alexander J Sundermann, Jenna Wiens, and Erica S Shenoy Leveraging electronic data to expand infection detection beyond traditional settings and definitions , Antimicrobial Stewardship & Healthcare Epidemiology, 3(1), 2023.
- Shengpu Tang and Jenna Wiens Counterfactual-Augmented Importance Sampling for Semi-Offline Policy Evaluation , Advances in Neural Information Processing Systems (NeurIPS), 2023. [CODE]
- Erkin Otles, Brian Denton and Jenna Wiens Updating Clinical Risk Stratification Models Using Rank-Based Compatibility: Approaches for Evaluating and Optimizing Clinician-Model Team Performance , Machine Learning for Healthcare Conference, 2023.
- Harry Rubin-Faclone, Joyce Lee, and Jenna Wiens Denoising Autoencoders for Learning from Noisy Patient-Reported Data , CHIL, 2023. [CODE]
- Donna Tjandra and Jenna Wiens Leveraging an Alignment Set in Tackling Instance-Dependent Label Noise , CHIL, 2023. [CODE]
- Harry Rubin-Faclone, Joyce Lee, and Jenna Wiens Forecasting with Sparse but Informative Variables: A Case Study in Predicting Blood Glucose , AAAI, 2023. [CODE]
- Aaman Rebello, Shengpu Tang, Jenna Wiens, Sonali Parbhoo Leveraging Factored Action Spaces for Off-Policy Evaluation , Presented at Workshop on Counterfactuals in Minds and Machines at ICML 2023 and FrontiersLCD Workshop at ICML 2023, July 2023. [CODE]
2022
- Safa Jabri, Wendy Carender, Jenna Wiens, and Kathleen Sienko Automatic ML-based vestibular gait classification: examining the effects of IMU placement and gait task selection , Journal of NeuroEngineering Rehabilitation 19, 132 (2022).
- Meghana Kamineni, Erkin Otles, Jeeheh Oh, Krishna Rao, Vincent B. Young, Benjamin Y Li, Lauren West, David Hooper, Erica Shenoy, John Guttag, Jenna Wiens and Maggie Makar Prospective evaluation of data-driven models to predict daily risk of Clostridioides difficile infection at two large academic health centers , Infection Control & Hospital Epidemiology, 1-4 2022.
- Harry Rubin-Falcone, Ian Fox, Emily Hirscheld, Lynn Ang, Rodica Pop-Busui, Joyce Lee and Jenna Wiens Association Between Management of Continuous Subcutaneous Basal Insulin Administration and HbA1C , Journal of Diabetes Science and Technology, 16(5), 1120-1127, 2022.
- Meera Krishnamoorthy, Piyush Ranjan, John R Erb-Downward, Robert P Dickson, and Jenna Wiens AMAISE: a machine learning approach to index-free sequence enrichment , Communications Biology, 5(1), 1-11, 2022. [CODE]
- Sarah Jabbour, David Fouhey, Ella Kazerooni, Jenna Wiens and Michael Sjoding Combining chest x-rays and EHR data using machine learning to diagnose acute respiratory failure , Journal of the American Medical Informatics Association, 2022. [CODE]
- Jenna Wiens, Melissa Creary, and Michael Sjoding AI Models in Healthcare are not Colorblind and We Should Not Be Either , The Lancet Digital Health, 2022.
- Donna Tjandra, Raymond Migrino, Bruno Giordani, and Jenna Wiens Use of blood pressure measurements extracted from the electronic health record in predicting Alzheimer’s disease: A retrospective cohort study at two medical centers , Alzheimer’s & Dementia, 2022. [CODE]
- Fahad Kamran, Shengpu Tang, Erkin Otles, Dustin S McEvoy, Sameh N Saleh, Jen Gong, Benjamin Y Li, Sayon Dutta, Xinran Liu, Richard J Medford, Thomas S Valley, Lauren R West, Karandeep Singh, Seth Blumberg, John P Donnelly, Erica S Shenoy, John Z Ayanian, Brahmajee K Nallamothu, Michael W Sjoding, and Jenna Wiens Early Identification of patients admitted to hospital for COVID-19 at risk of clinical deterioration: model development and multisite external validation study , British Medical Journal 376, 2022. [CODE]
- Emily Mu, Sarah Jabbour, Adrian V Dalca, John Guttag, Jenna Wiens, Michael W Sjoding Augmenting existing deterioration indices with chest radiographs to predict clinical deterioration , PloS one 17 (2), 2022.
- Kayte Spector-Bagdady, Shengpu Tang, Sarah Jabbour, Nicholson Price, Ana Bracic, Sachin Kheterpal, Chad M. Brummett, and Jenna Wiens Respecting Autonomy And Enabling Diversity: The Effect Of Eligibility And Enrollment On Research Data Demographics , Health Affairs 40 (12) 1892-1899, 2022.
- Jiaxuan Wang, Sarah Jabbour, Maggie Makar, Michael Sjoding, and Jenna Wiens Learning Concept Credible Models for Mitigating Shortcuts , NeurIPS, 2022.
- Shengpu Tang, Maggie Makar, Michael Sjoding, Finale Doshi-Velez and Jenna Wiens Leveraging Factored Action Spaces for Efficient Offline Reinforcement Learning in Healthcare , NeurIPS, 2022. [CODE]
- Trenton Chang, Michael Sjoding, and Jenna Wiens Disparate Censorship & Undertesting: A Source of Label Bias in Clinical Machine Learning , MLHC, 2022. [CODE]
- Harry Rubin-Falcone, Joyce Lee, and Jenna Wiens Forecasting with Sparse but Informative Variables: A Case Study in Predicting Blood Glucose , KDD Workshop, 2022.
2021
- Fahad Kamran, Katherine Harrold, Jonathan Zwier, Wendy Carender, Tian Bao, Kathleen Sienko, and Jenna Wiens Automatically evaluating balance using machine learning and data from a single inertial measurement unit , Journal of NeuroEngineering and Rehabilitation 18 (1), 1-7, 2021.
- Jeremiah Hauth, Safa Jabri, Fahad Kamran, Eyoel W Feleke, Kal Niguisie, Lauro V Ojeda, Shirley Handelzalts, Linda Nyquist, Neil B Alexander, Xun Huan, Jenna Wiens, and Kathleen Sienko Automated loss-of-balance event identification in older adults at risk of falls during real-world walking using wearable inertial measurement units , Sensors 21 (14), 2021.
- Fahad Kamran, Victor C Le, Adam Frischknecht, Jenna Wiens, and Kathleen Sienko Non-invasive Estimation of Hydration Status in Athletes Using Wearable Sensors and a Data-Driven Approach based on Orthostatic Changes , Sensors 21 (13), 2021.
- Begum Topcuoglu, Zena Lapp, Kelly Sovacool, Evan Snitkin, Jenna Wiens, and Patrick Schloss mikropml: user-friendly R package for supervised machine learning pipelines , Journal of Open Source Software (6) 61 2021.
- Saige Rutherford, Pascal Sturmfels, Mike Angstadt, Jasmine Hect, Jenna Wiens, Marion I van en Heuvel, Dustin Scheinost, Chandra Sripada, and Moriah Thomason Automated Brain Masking of Fetal Functional MRI with Open Data , Neuroinformatics, 1-13, 2021.
- Zena Lapp, Ebbing Lautenbach, Jennifer Han, Jenna Wiens, Ellie Goldstein, and Evan Snitken Patient and microbial genomic factors associated with carbapenem-resistant Kelbsiella pneumoniae extraintestinal colonization and infection , mSystems 2021.
- Jaewon Hur, Shengpu Tang, Vidhya Gunaseelan, Joceline Vu, Chad M. Brummett, Michael Englesbe, Jennifer Waljee and Jenna Wiens Predicting Postoperative Opioid Use with Machine Learning and Insurance Claims in Opioid-Naive Patients , the American Journal of Surgery, 2021. [CODE]
- Karandeep Singh, Thomas S. Valley, Shengpu Tang, Benjamin Y. Li, Fahad Karman, Michael W. Sjoding, Jenna Wiens, Erkin Otles, John P. Donnelly, Melissa Y. Wei, Jonathon P. McBride, Jie Cao, Carleen Penoza, John Z. Ayanian, and Brahmajee K. Nallamothu Validating a Widely Implemented Deterioration Index Model Among Hospitalized COVID-19 Patients , Annals of the American Thoracic Society 2021.
- Joel Castellanos, Cheng Perng Phoo, James Eckner, Lea Franco, Steve Broglio, Mike McCrea, Thomas McAllister, and Jenna Wiens Predicting Risk of Sport-Related Concussion in Collegiate Athletes and Military Cadets: A Machine Learning Approach using Baseline Data from the CARE Consortium Study , Sports Medicine, 2021, pg. 1-13.
- Erkin Otles, Jeeheh Oh, Benjamin Li, Michelle Bochinsky, Hyeon Joo, Justin Ortwine, Erica Shenoy, Laraine Washer, Vincent Young, Krishna Rao and Jenna Wiens Mind the Performance Gap: Examining Dataset Shift During Prospective Validation , MLHC, August 2021.
- Shengpu Tang and Jenna Wiens Model Selection for Offline Reinforcement Learning: Practical Considerations for Healthcare Settings , MLHC, August 2021. [CODE]
- Jiaxuan Wang, Jenna Wiens, and Scott Lundberg Shapley Flow: A Graph-based Approach to Interpreting Model Predictions , AISTATS, April 2021. [CODE]
- Donna Tjandra, Yifei He, and Jenna Wiens A Hierarchical Approach to Multi-Event Survival Analysis , AAAI, February 2021. [CODE]
- Fahad Kamran and Jenna Wiens Estimating Calibrated Individualized Survival Curves with Deep Learning , AAAI, February 2021. [CODE]
- Emily Mu, Sarah Jabbour, Adrian Dalca, John Guttag, Jenna Wiens, and Michael Sjoding Joint Modeling of EHR and CXR Data to Predict COVID-19 Deterioration , Open Forum Infectious Diseases, 8 (Supplement 1), 2021.
- Erkin Otles, Jeeheh Oh, Alieysa Patel, Micah Keidan, Vincent B Young, Krishna Rao, and Jenna Wiens Comparative Assessment of a Machine Learning Model and Rectal Swab Surveillance to Predict Hospital Onset Clostridioides difficile , Open Forum Infectious Diseases, 8 (Supplement 1), 2021.
- Jeeheh Oh and Jenna Wiens A Data-Driven Approach to Estimating Infectious Disease Transmission from Graphs: A Case of Class Imbalance Driven Low Homophily , ACM CHIL Workshops, online, April 2021.
2020
- Karandeep Singh, Thomas S. Valley, Shengpu Tang, Benjamin Y. Li, Fahad Kamran, Michael W. Sjoding, Jenna Wiens, Erkin Otles, John P. Donnelly, Melissa Y. Wei, Jonathon P. McBride, Jie Cao, Carleen Penoza, John Z. Ayanian, Brahmajee K. Nallamothu Evaluating a Widely Implemented Proprietary Deterioration Index Model Among Hospitalized COVID-19 Patients , Annals of the American Thoracic Society, 2020. [CODE]
- Shengpu Tang, Parmida Davarmanesh, Yanmeng Song, Danai Koutra, Michael W. Sjoding, and Jenna Wiens Democratizing EHR Analysis with FIDDLE: A Flexible Preprocessing Pipeline for Structured Clinical Data , Journal of American Medical Informatics Association, 2020. [CODE]
- Michael Fenstermaker, Scott A Tomlins, Karandeep Singh, Jenna Wiens, and Todd M Morgan Development and Validation of a Deep-Learning Model to Assist with Renal Cell Carcinoma Histopathologic Interpretation , Urology, July 2020.
- Begum Topcuoglu, Nicholas Lesniak, Mack Ruffin, Jenna Wiens, and Patrick Schloss A Framework for Effective Application of Machine Learning to Microbiome-Based Classification Problems , mBio, June 2020.
- Donna Tjandra, Raymond Migrino, Bruno Giordani, and Jenna Wiens Cohort Discovery and Risk Stratification for AD: An EHR-based Approach , Alzheimer’s & Dementia: Translational Research & Clinical Interventions 6, no. 1: e12035, 2020. [CODE]
- Shengpu Tang, Grant T Chappell, Amanda Mazzoli, Muneesh Tewari, Sung Won Choi, Jenna Wiens Predicting Acute Graft-versus-Host Disease Using Machine Learning and Longitudinal Vital Sign Data from Electronic Health Records , JCO Clinical Cancer Informatics, February 2020. [CODE]
- Vincent X Liu, Jenna Wiens ‘No growth to date’? Predicting positive blood cultures in critical illness , Intensive Care Medicine, January 2020.
- Jenna Wiens, W. Nicholson Price II, and Michael W. Sjoding Diagnosing bias in data-driven algorithms for healthcare , Nature Medicine, January 2020.
- Sarah Jabbour, David Fouhey, Ella Kazerooni, Michael W. Sjoding, and Jenna Wiens Deep Learning Applied to Chest X-Rays: Exploiting and Preventing Shortcuts , MLHC, August 2020. [CODE] [VIDEO]
- Ian Fox, Joyce Lee, Rodica Pop-Busui, and Jenna Wiens Deep Reinforcement Learning for Closed-Loop Blood Glucose Control , MLHC, August 2020. [CODE] [VIDEO]
- Shengpu Tang, Aditya Modi, Michael W. Sjoding, and Jenna Wiens Clinician-in-the-Loop Decision Making: Reinforcement Learning with Near-Optimal Set-Valued Policies , ICML, July 2020. [CODE] [VIDEO]
- Harry Rubin-Falcone, Ian Fox, Jenna Wiens Deep Residual Time-Series Forecasting: Application to Blood Glucose Prediction , KDH (Knowledge Discovery in Healthcare Data), August 2020. [CODE] [VIDEO]
2019
- Ian Fox, and Jenna Wiens Advocacy Learning: Learning through Competition and Class-Conditional Representations , IJCAI , 2019. [CODE]
- Jeeheh Oh, Jiaxuan Wang, Shengpu Tang, Michael Sjoding, and Jenna Wiens, Relaxed Weight Sharing: Effectively Modeling Time-Varying Relationships in Clinical Time-Series , MLHC , 2019. [CODE]
- Michael Sjoding, Shengpu Tang, Parmida Davarmanesh, Yanmeng Song, Danai Koutra, Jenna Wiens. et al., Democratizing EHR Analyses a Comprehensive Pipeline for Learning from Clinical Data , MLHC (Clinical Abstract) , 2019 [to appear]. [CODE]
- Erkin Otles, Haozhu Wang, et al., Return to Work After Injury: A Sequential Prediction and Prescription Problem , MLHC (Clinical Abstract) , 2019 [to appear].
- Donna Tjandra, Raymond Migrino, Bruno Giordani, and Jenna Wiens, An EHR-based Cohort Discovery Tool for Identifying Probable AD , Alzheimer's Association International Conference (AAIC) , 2019.
- Donna Tjandra, Raymond Migrino, Bruno Giordani, and Jenna Wiens, EHR-based Patient Risk Stratification Tool for Probable AD , Alzheimer's Association International Conference (AAIC) , 2019.
- Ian Fox and Jenna Wiens, Reinforcement Learning for Blood Glucose Control: Challenges and Opportunities , RL4RealLife Workshop, ICML , 2019.
- Ben Li, Jeeheh Oh, Vincent Young, Krishna Rao, and Jenna Wiens, Using Machine Learning and the Electronic Health Record to Predict Complicated Clostridium difficile Infection , Open Forum Infectious Diseases 6(5) , 2019. [CODE]
- Daniel Zeiberg, Tejas Prahlad, Brahmajee Nallamothu, Theordore J. Iwashyna, Jenna Wiens* and Michael Sjoding* , Machine learning for patient risk stratification for acute respiratory distress syndrome , PLOS ONE 14 (13) , 2019. *co-senior authors [CODE]
- Tian Bao, Brooke N. Klatt, Susan L. Whitney, Kathleen H. Sienko, and Jenna Wiens Automatically evaluating balance: a machine learning approach , IEEE Transactions on Neural Systems and Rehabilitation Engineering , 2019.
- Saige Rutherford, Pascal Sturmfels et al., Observing the origins of human brain development: Automated processing of fetal fMRI , bioRxiv , 2019.
2018
- Devendra Goyal, Donna Tjandra, et al., Characterizing heterogeneity in the progression of Alzheimer's disease using longitudinal clinical and neuroimaging biomarkers , Alzheimer's &Dementia: Diagnosis, Assessment &Disease Monitoring , 2018. [CODE]
- Jeeheh Oh, Jiaxuan Wang, and Jenna Wiens Learning to Exploit Invariances in Clinical Time-Series Data using Sequence Transformer Networks , MLHC , 2018. [CODE]
- Pascal Sturmfels et al., A Domain Guided CNN Architecture for Predicting Age from Structural Brain Images , MLHC , 2018.
- Jenna Wiens and James Fackler, Striking the Right Balance - Applying Machine Learning to Pediatric Critical Care Data , Pediatric Critical Care Medicine , 2018.
- Ian Fox, Lynn Ang, Mamta Jaiswal, Rodica Pop-Busui, Jenna Wienset al., Deep Multi-Output Forecasting: Learning to Accurately Predict Blood Glucose Trajectories , KDD , August 2018. [CODE]
- Jiaxuan Wang, Jeeheh Oh, Haozhu Wang, Jenna Wienset al., Learning Credible Models , KDD , August 2018. [CODE]
- Jeeheh Oh, Maggie Makar et al., A Generalizable, Data-Driven Approach to Predict Daily Risk of Clostridium difficile Infection at Two Large Academic Health Centers , Infection Control and Hospital Epidemiology , March 2018.
- Devendra Goyal, Zeeshan Syed, and Jenna Wiens, Clinically Meaningful Comparisons Over Time: An Approach to Measuring Patient Similarity based on Subsequence Alignment , arXiv:1803.00744 , 2018.
- Jiaxuan Wang, Ian Fox, Jonathan Skaza, Nick Linck, Satinder Singh, and Jenna Wiens, The Advantage of Doubling: A Deep Reinforcement Learning Approach to Studying the Double Team in the NBA , Sloan Sports Analytics Conference , February 2018. [Poster ]
- Maggie Makar, John Guttag, and Jenna Wiens, Learning the Probability of Activation in the Presence of Latent Spreaders , AAAI , February 2018. (oral presentation)
2017
- Jenna Wiens, Graham Snyder et al., Potential Adverse Effects of Broad-Spectrum Antimicrobial Exposure in the Intensive Care Unit , Open Forum Infectious Diseases , December 2017.
- Eli Sherman et al., Leveraging Clinical Time-Series Data for Prediction: A Cautionary Tale , AMIA Annual Symposium , November 2017. (oral presentation)
- Jeeheh Oh, Maggie Makar et al., A Data-Driven Approach to Predict Daily risk of Clostridium difficile Infection at Two Large Academic Health Centers , Infectious Disease Week , October 2017.
- Jenna Wiens and Erica Shenoy, Machine Learning for Healthcare: On the Verge of a Major Shift in Healthcare Epidemiology , Clinical Infectious Diseases , August 2017.
- Ian Fox et al., Contextual Motifs: Increasing the Utility of Motifs using Contextual Data , KDD , August 2017. (Oral Presentation Acceptance Rate: 8.5%)
2016 - 2011
- Jose Javier Gonzalez Ortiz, Cheng Perng Phoo, and Jenna Wiens, Heart Sound Classification Based on Temporal Alignment Techniques , Computing in Cardiology , September 2016.[Code: CODElink ]
- Mason Wright and Jenna Wiens, Method to their March Madness: Insights from Mining a Novel Large-Scale Dataset of Pool Brackets , KDD Workshop on Large-Scale Sports Analytics , August 2016.
- Jenna Wiens, John Guttag, and Eric Horvitz, Patient Risk Stratification with Time-Varying Parameters: A Multitask Learning Approach , JMLR , April 2016.
- Avery McIntyre et al., Recognizing and Analyzing Ball Screen Defense in the NBA , Sloan Sports Analytics Conference , March 2016.[Slides: pdf ]
- Abhishek Bafna and Jenna Wiens, Automated Feature Learning: Mining Unstructured Data for Useful Abstractions , ICDM , November 2015.
- Abhishek Bafna and Jenna Wiens, Learning Useful Abstractions from the Web , AMIA Annual Symposium , November 2015. (poster)
- Sai R. Gouravajhala et al., An LED Blink is Worth a Thousand Packets: Inferring a Networked Device's Activity from its LED Blinks , USENIX Summit on Information Technologies for Health , August 2015.
- Devendra Goyal, Zeeshan Syed, and Jenna Wiens, Predicting Disease Progression in Alzheimer's Disease , MUCMD , August 2015.
- Jenna Wiens et al., Learning Data-Driven Patient Risk Stratification Models for Clostridium difficile ,Open Forum Infectious Diseases , July 2014.
- Jenna Wiens, Learning to Prevent Healthcare-Associated Infections: Leveraging Data Across Time and Space to Improve Local Predictions, PhD Thesis, MIT, May 2014.
- Jenna Wiens et al., Automatically Recognizing On-Ball Screens , Sloan Sports Analytics Conference , Feb 2014.
- Jenna Wiens et al., A Study in Transfer Learning: Leveraging Data from Multiple Hospitals to Enhance Hospital-Specific Predictions , Journal of the American Medical Informatics Association , Jan 2014.
- Jenna Wiens et al., To Crash or Not to Crash: A quantitative look a the relationship between offensive rebounding and transition defense in the NBA , Sloan Sports Analytics Conference , March 2013.
- Jenna Wiens et al., Patient Risk Stratification for Hospital-Associated C. diff as a Time-Series Classification Task , Neural Information Processing Systems (NeurIPSNIPS) , Dec 2012. [Video ]
- Jenna Wiens et al., Learning Evolving Patient Risk Processes for C. diff Colonization , ICML Workshop on Clinical Data Analysis , June 2012.[Slides: pdf ]
- Jenna Wiens et al., On the Promise of Topic Models for Abstracting Complex Medical Data: A Study of Patients and their Medications , NeurIPSNIPS Workshop on Personalized Medicine , December 2011.
- Jenna Wiens and John Guttag, Patient-Specific Ventricular Beat Classification without Patient-Specific Expert Knowledge: A Transfer Learning Approach , IEEE EMBS Conference , September 2011.
- Jenna Wiens and John Guttag, Active Learning Applied to Patient-Adaptive Heartbeat Classification , Neural Information Processing Systems (NeurIPSNIPS) , December 2010.
- Jenna Wiens, Machine Learning for Ectopic Beat Classification , Master's thesis, MIT, May 2010.
- Jenna Wiens and John Guttag, Patient-Adaptive Ectopic Beat Classification using Active Learning . , Computing in Cardiology (CinC) September 2010. [Slides: pptx ]
Multimedia
Women in Tech Shown | Scientific American | ACP Hospitalist Article | MIT Tech Review Article | SSAC16 | Invited Talk at Wellesley College: Big Data's Impact in Medicine, Finance, and Sports | SSAC13: To Crash or not to Crash | ESPN TrueHoop TV: Interview with Henry Abbott | ESPN TrueHoop: Commentary | Grantland Interview | NeurIPSNIPS 2012 Spotlight | NeurIPSNIPS Workshops 2011 Spotlight