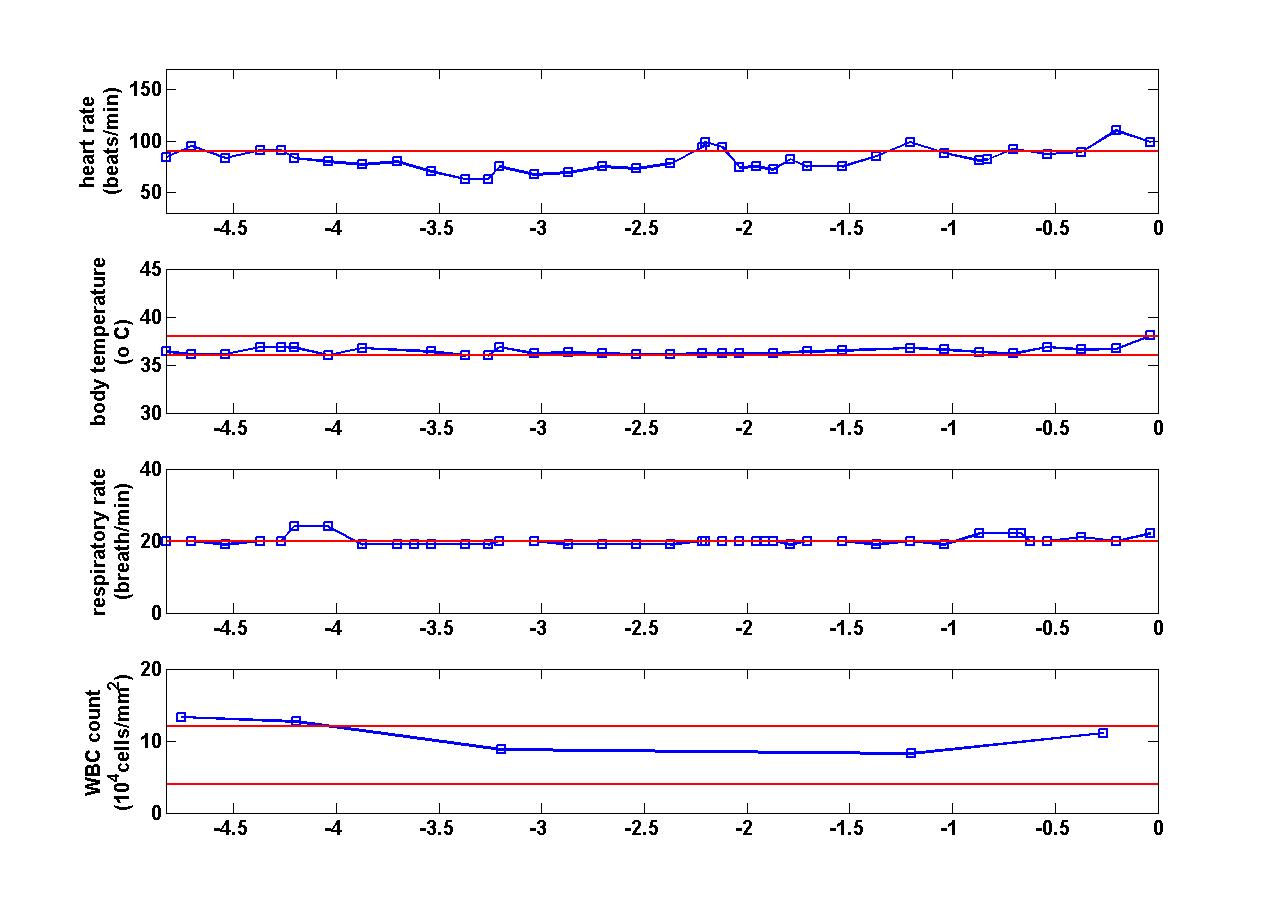
A major challenge in modern mental health science is to quantify the neural mechanisms that 1) distinguish patients with various mental health disorders from healthy controls, and 2) characterize the effects of various psychoactive drugs. Toward this end, this project applies machine learning methodology (e.g., embedded feature selection, multi-task learning) to functional neuroimaging (fMRI) data across a variety of mental health disorders. For each mental health disorder or drug analyzed, the goal is to infer a connectome, which is a network of pairs of brain regions whose functional correlation differs for the given treatment/condition relative to baseline. A single study may involve data from several scanners, with multiple subjects per scanner, while a single subject's brain scan involves millions of voxels. Thus, scalability and scanner- and patient-induced variability are major challenges in this work.
Collaborator: Chandra Sripada, UM Depts of
Psychiatry and Philosophy
Graduate student: Takanori Watanabe
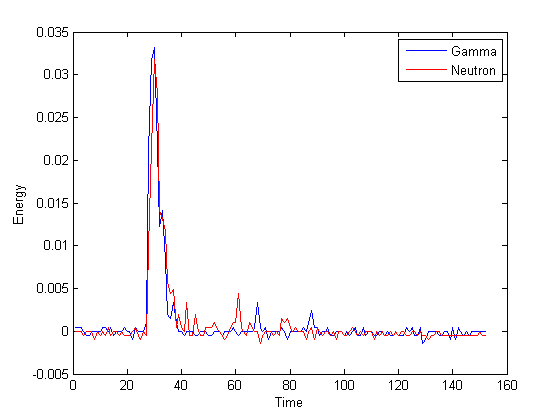
Illicit nuclear material is characterized by the energy distribution (spectrum) of emitted neutrons. Organic scintillation detectors are an important technology for neutron detection, owing to their efficiency and cost. These detectors convert the energy deposited by nuclear particles into light, which is then converted to an electrical waveform and digitally sampled, yielding a pulse-shaped signal. Since organic scintillation detectors are also sensitive to gamma-rays, it is necessary to classify neutron and gamma-ray pulses. To design a classifier we employ training data gathered from known gamma-ray and neutron sources. However, the application of classification methodology is unconventional in two respects. 1) Pure data sets for each particle type are unavailable, owing to contamination from background radiation, and other factors; this leads to the problem of classification with label noise. 2) In field applications, measurements are likely to have different energy distributions compared with training data, owing to unknown source material and shielding; this is a new instance of a problem known in machine learning circles as domain adaptation. This project is developing new methodology and theory for these two challenges. Scalability is also a major concern for two reasons: First, training data sets may contain millions of examples, and second, trained classifiers must have efficient implementations in hardware, given nanosecond sampling rates combined with thousands of measured events per second.
Collaborators: Sara Pozzi and Marek Flaska, UM Dept. of Nuclear
Engineering and Radiological Sciences, and David Wentzloff,
EECS.
Graduate students: Tyler Sanderson, Gregory
Handy
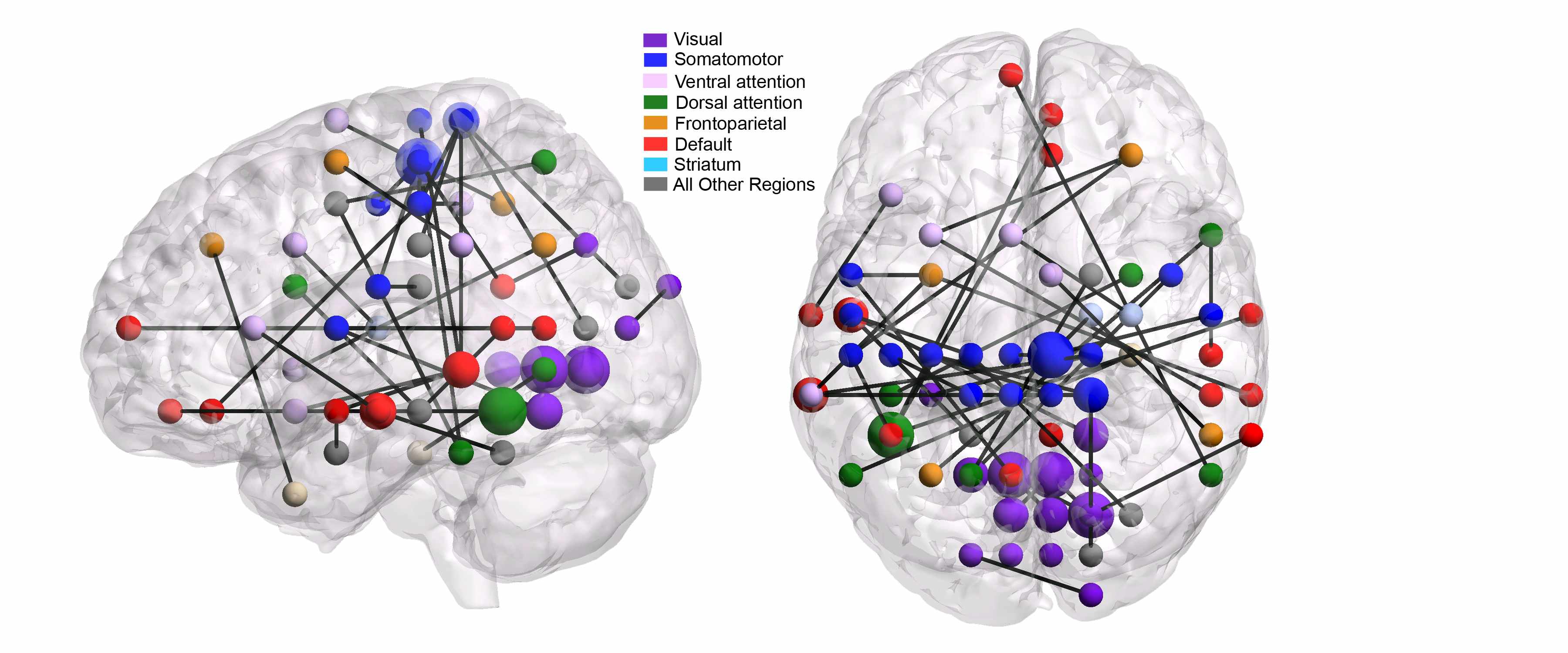
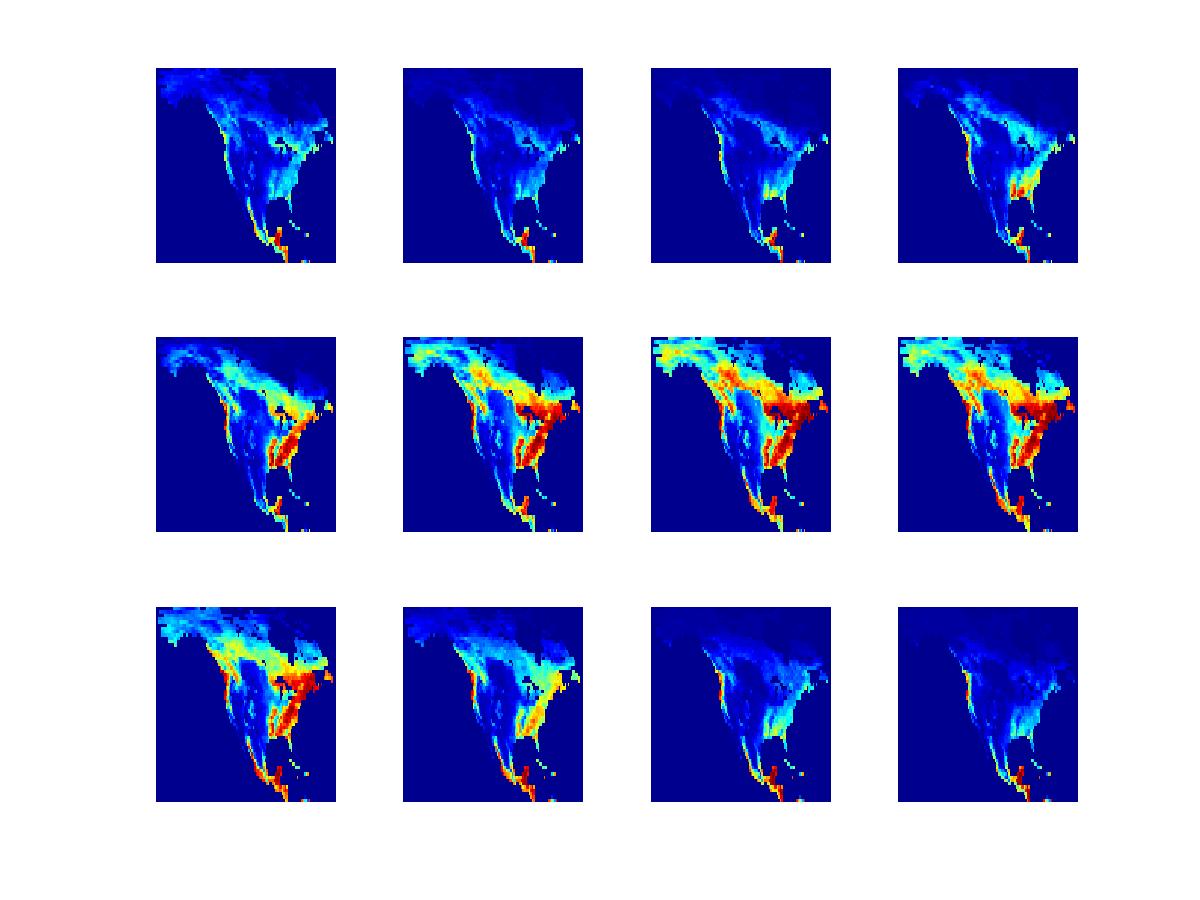
Through ground- or satellite-based measurements, various environmental variables related to carbon dioxide uptake can be measured or computed across a broad spatial and temporal range. Examples of such variables include leaf area index and carbon dioxide flux. Climate scientists have a keen interest in detecting anomalies in these spatio-temporal data sets. As with most anomaly detection problems, a major challenge is the lack of examples of known anomalies, or even examples of known non-anomalies. However, domain experts can provide certain qualitative descriptions of normal and anomalous trends, together with estimates of spatio-temporal covariance. Our approach has been to translate these descriptions into quantitative low-rank/sparsity assumptions, leading to tractable inference strategies with interpretable outputs.
Collaborators: Long Nguyen, UM Dept. of Statistics, and Anna Michilak and
Vineet Yadav, Carnegie Institution for Science.
Graduate students: Hossein
Keshavarz, Benjamin Schwartz
Support: NSF
In many biomedical domains, there is a need to design classifiers that are
specifically adapted to each new patient. For example, in ECG data, there is a need
to label each heartbeat as normal or abnormal, while in EEG data, it is desirable to
label a temporal window according to whether it precedes a seizure. In both
cases, the definition of normal and abnormal will vary from patient to patient. Even
if labeled examples are acquired for many training patients, this information must
still be adapted to each new patient to improve classification accuracy. In ongoing
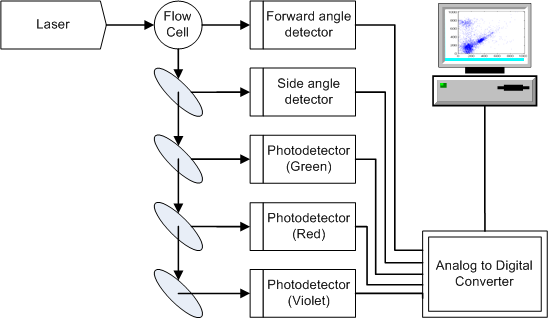
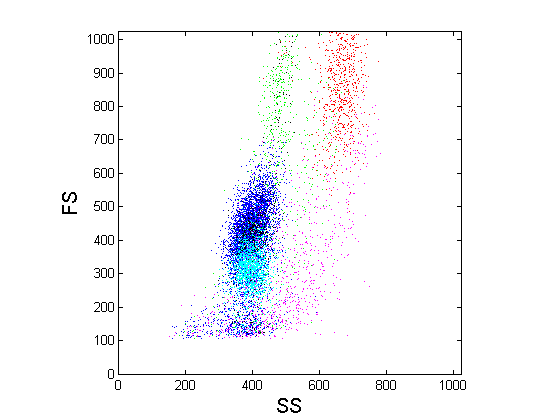
work, we have developed this idea in the context of flow cytometry. Flow cytometry is
a kind of high-throughput biological assay that is capable of quantifying physical
and chemical properties of individual cells, and is used by pathologists to diagnose
and classify a variety of blood-related disorders, including leukemias and
lymphomas. A problem of interest to pathologists is to
automatically classify cells according to type (lymphocyte, granulocyte, etc.). In
principle, it is possible to design such classifiers by training on labeled data
sets; however, because of patient-to-patient variability, there is a need to design
patient-specific classifiers that adapt to the cell distribution of each patient.
Patient-specific classification is a kind of problem known as transfer learning in
machine learning; we have developed a general-purpose solution to this problem that
is potentially applicable in a variety of application domains, such as those
mentioned above. Furthermore, flow cytometry data sets may contain hundreds of
thousands of cells, and therefore scalable algorithms are
necessary.
|
|
Collaborators:: Gilles Blanchard, University of Potsdam, and Drs. William Finn and Lloyd Stoolman, UM Dept. of Pathology
Graduate student: Gyemin Lee
Support: NSF, UM
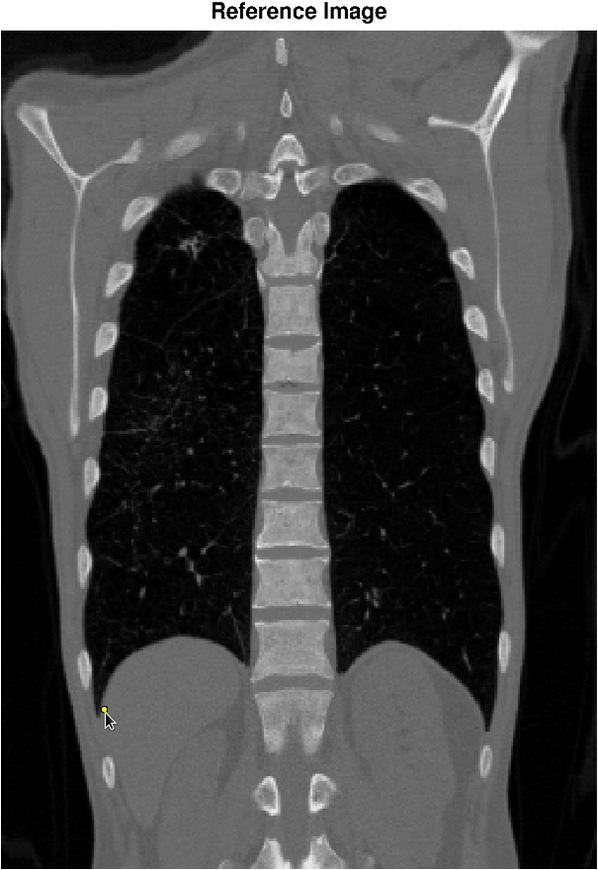
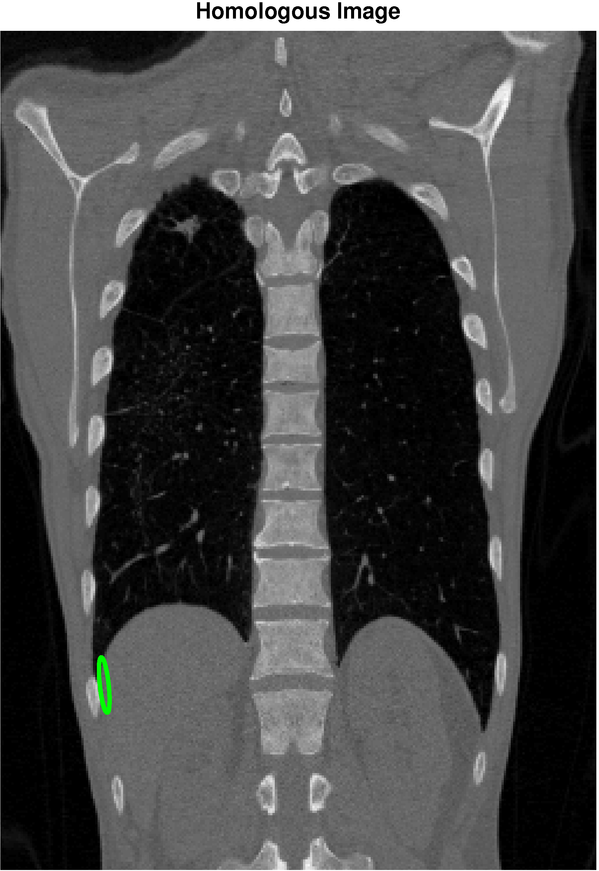
Image registration is the task of finding a spatial transformation that brings two images into alignment. It is a critical step in medical image analysis, for example, to align images of a patient taken under different modalities such at MRI and CT. Our objective is to apply machine learning to quantify the uncertainty in image registration algorithms. The figure below shows a reference image (left) and a holomogous image (right) that has been registered to the reference image. The selected point in the reference image corresponds, with high confidence, to a point in the region shown in the homologous image. In this example, the orientation of the confidence region reflects uncertainty due to the sliding motion of the diaphragm.
 |  |
Collaborators: Charles Meyer, UM Dept. of Radiology, and Alfred Hero, EECS.
Graduate student: Takanori Watanabe
Support: NIH
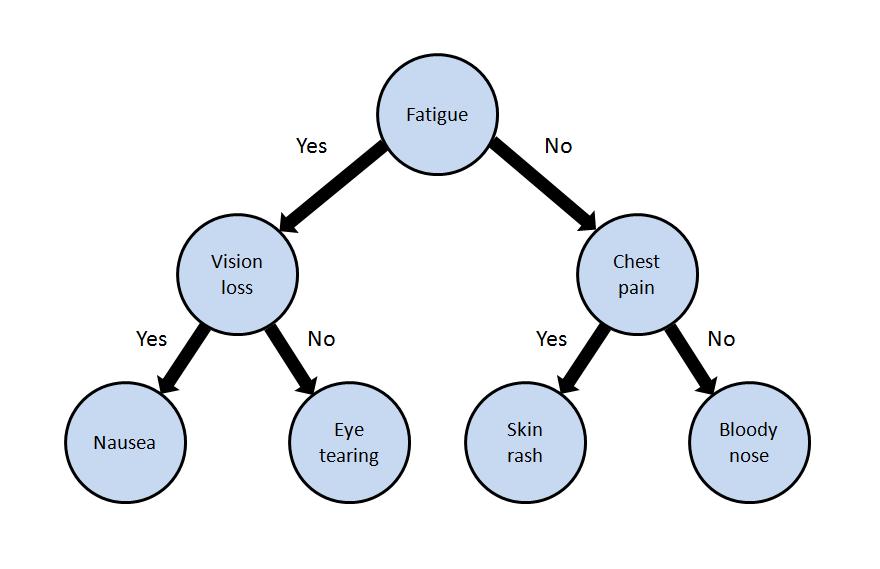
When first responders arrive on the scene of an accident or attack involving toxic chemicals, they need to rapidly identify the chemical. One way they do this is by observing which symptoms are expressed in victims. Our work centers on designing decision trees that guide first responders to test those symptoms that will most rapidly lead to chemical identification. We have developed algorithms and user interfaces that are designed to be both comprehensible to first responders, and robust to noise and uncertainty in the data and environment.
|
|
Graduate student: Gowtham Bellala
Support: NSF, UM, CDC/NIOSH